Summary
Our lab’s primary focus is on understanding microalgae community diversity and evolution over time, in both marine and alpine environments. We ultimately hope to understand how these communities functionally adapt to changing environments, especially as our marine and alpine environments in the pacific northwest are changing rapidly. We use high-throughput sequencing of DNA or RNA to characterize the taxonomic diversity (large scale tag sequencing projects) and functional diversity (metagenomics).
Diversity and Dynamics of Snow Algae Communities
Pink snow, commonly referred to as watermelon snow, represents a community of organisms that include microalgae and bacteria. Though the algae in these communities have been known a long time from microscope studies, ours is the first large-scale DNA survey of the whole community using the newest technology in high-throughput DNA sequencing. Using these new powerful environmental genomic techniques it work in a number of areas: (1) are there are biogeographic patterns to snow algae communities, (2) how does the algae community diversity change with elevation and latitude, (3) how may these communities be affected by a reduction of snowpack in the future, (4) population-level diversity in snow algae blooms, and (5) snow algae lifecycles and physiology.

See News for the latest updates on our work in the media, and Alpine Algae Research Blog that gives updates on our past climbing and skiing sampling adventures .
We also run The Living Snow Project: a citizen science/community science/climber and skier science program to engage the public in helping us sample for snow algae.
Phytoplankton Community Structure Across Temporal and Spatial Scales in Bellingham Bay

We have been working on characterizing the whole phytoplankton community over space and time through periodic summer hypoxic conditions in Bellingham Bay over the past 4 years. We characterize the various size fractions of phytoplankton via flow cytometry, microscopy, and quantitative cell counts, and will be doing high-throughput surveys of 18S SSU ribosomal genes (V4 region).
This year, we partnered with the Bellingham Community Boating Center to characterize bioluminescent plankton in Bellingham Bay, collected on kayak tours. Robin talks about this phenomenon and the organisms that produce bioluminescence in a piece we made with Oregon Field Guide.
Phycomate Prasinophyte Diversity in the Salish Sea
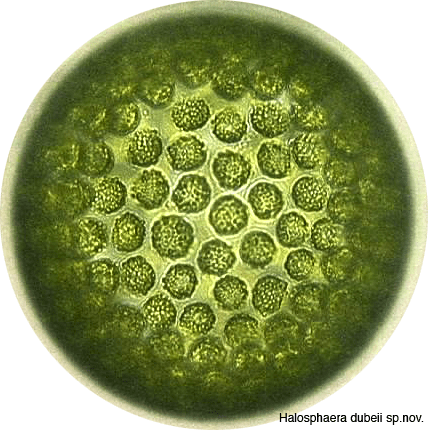
In 2006, while in graduate school, Dr. Kodner came to Washington for the first time to collect Halosphaera, a genus of phycomate prasinophyte that cannot be cultured and is difficult to find. It is our best modern analog for the most ancient eukaryotic fossils and thus, an important evolutionary lineage. We have collected samples in 2013 and 2014 to begin to characterize this population using more advanced microscopy techniques and genomics.
Improving phylogenetic-based metagenome analysis pipelines
We have worked in the past on optimizing bioinformatics methods for environmental microeukaryote populations. This includes helping develop the pplacer software package with Erick Matsen at the Fred Hutchenson Cancer Research Center. Since its publication in 2010, we have been working on a pipeline to run pplacer across thousands of genes in any large sequence data set.